1+2*(3+5)[1] 17By the end of this class you will
glimpse(), names(), nrow(), ncol(), count()ggplot()We will do this using water lead content data from Flint, MI. The following paragraph will be useful in evaluating the lead amount values we’ll see in the dataset.
While there is no completely safe amount of lead consumption, the limit allowed by the Lead and Copper Rule (LCR) of 1991 is 15 parts per billion (ppb). If this is exceeded in more than 10% of homes tested (or if the 90th percentile value of the total sample is above 15 ppb), action is required. And to make sure problems are caught, sampling for lead in water is supposed to target the “worst-case” homes – those in areas served by lead pipes.
Files, plots, viewer, environment, etc. panes
Console
Editor
Writing code in the console
Basic math with R
1+2*(3+5)[1] 17Creating variables in R, the assignment operator (<-), and the Environment pane
age <- 27
age + 4[1] 31age + 9[1] 36R functions and packages and the Packages pane
Getting help with R and the Help pane
YAML: Metadata
Narrative: Edited with the visual editor (or the source editor)
Code: In code chunks
Chunk options (following #|)
Comments (following #)
Code
Running individual code chunks vs. rendering a document
We’ll use the tidyverse package for analysis, which offers functionality for data import, wrangling, visualization, and more.
# load a package
library(tidyverse)── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.4
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 3.4.4 ✔ tibble 3.2.1
✔ lubridate 1.9.2 ✔ tidyr 1.3.0
✔ purrr 1.0.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ purrr::%||%() masks base::%||%()
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorsLoading this package prints out a message. What does this message mean? How can we suppress the message from the output?
The read_csv() function can be used for reading CSV (comma separated values) files. The file we’re reading is called flint with the suffix (.csv) which indicates its file type.
flint <- read_csv("flint.csv")Rows: 813 Columns: 5
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (1): draw
dbl (4): id, zip, ward, lead
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.view(flint)One of two things may have happened:
flint in your Environment pane.Error in read_csv("flint.csv") : could not find function "read_csv".If (1) happened, great!
If (2) happened, let’s troubleshoot first before continuing.
The following variables are in the flint data frame:
id: sample ID number (identifies the home)zip: ZIP code in Flint of the sample’s locationward: ward in Flint of the sample’s locationdraw: which time point the water was sampled fromlead: lead content in parts per billion (ppb)We want to learn about the population using a sample.
In the case we want to learn about the lead content in all of Flint, MI homes but only have available water readings from a sample of homes (our data set).
Exercise 1: Look at the data, how many observations are there? How many variables?
nrow(flint)[1] 813ncol(flint)[1] 5There are 813 observations and 5 variables.
Let’s count() to find the number of different time points water was sampled with the count() function.
The first argument is flint: the data frame
The second argument is draw: the variable
count(flint, draw)# A tibble: 3 × 2
draw n
<chr> <int>
1 first 271
2 second 271
3 third 271We can achieve the same result with the following “piped” operation as well.
The first line is flint: the data frame
Then the pipe operator, read as “and then”, which places what comes before it as the first argument of what comes after it
The second line is count(draw)
flint |>
count(draw)# A tibble: 3 × 2
draw n
<chr> <int>
1 first 271
2 second 271
3 third 271We can use a similar approach to fund out how many unique homes are in the data set:
flint |>
count(id)# A tibble: 269 × 2
id n
<dbl> <int>
1 1 3
2 2 3
3 4 3
4 5 3
5 6 3
6 7 3
7 8 3
8 9 3
9 12 3
10 13 3
# ℹ 259 more rowsExercise 2: How many samples were taken from each zip code?
flint |>
count(zip)# A tibble: 8 × 2
zip n
<dbl> <int>
1 48502 3
2 48503 207
3 48504 165
4 48505 144
5 48506 132
6 48507 153
7 48529 3
8 48532 6Exercise 3: Which ZIP code had the most samples drawn? Hint: See the help for count.
flint |>
count(zip, sort = TRUE)# A tibble: 8 × 2
zip n
<dbl> <int>
1 48503 207
2 48504 165
3 48507 153
4 48505 144
5 48506 132
6 48532 6
7 48502 3
8 48529 3What is a statistic? It’s any mathematical function of the data. Sometimes, a statistic is referred to as “sample statistic” or “summary statistic”, since you compute it from a sample (the observed data) and not the entire population.
… and any other arbitrary function of the data you can come up with!
Exercise 4: Compute each of these statistics for lead ppb.
summary_table <- flint |>
summarize(mean_lead = mean(lead),
median_lead = median(lead),
var_lead = var(lead),
sd_lead = sd(lead))
summary_table# A tibble: 1 × 4
mean_lead median_lead var_lead sd_lead
<dbl> <dbl> <dbl> <dbl>
1 8.20 1.85 1718. 41.5Let’s take a look at the distribution of lead content in homes in Flint, MI.
ggplot(flint, aes(x = lead)) +
geom_histogram()`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.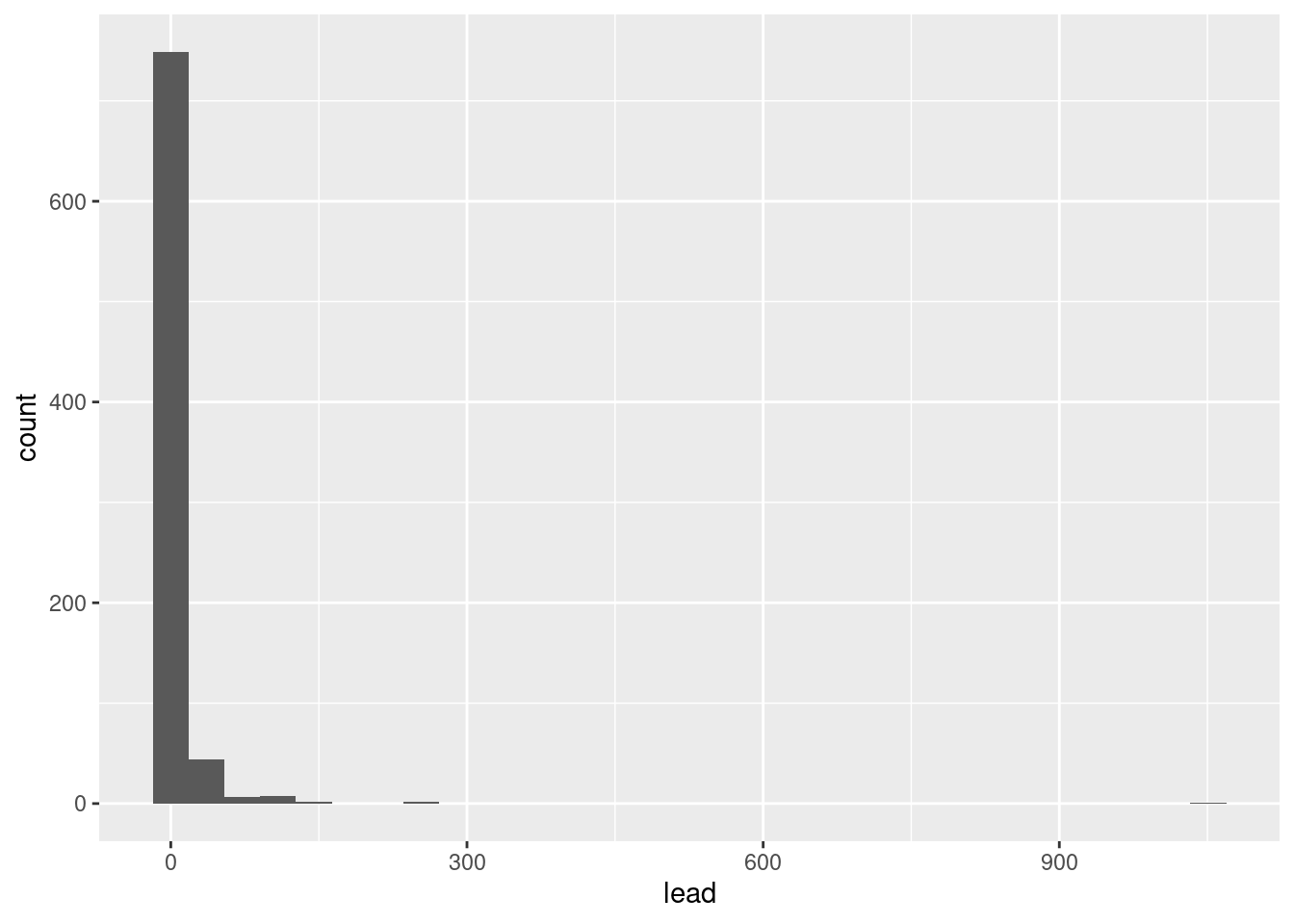
We can make this plot look nicer/more useful by adjusting the number of bins and zooming into the x-axis.
ggplot(flint, aes(x = lead)) +
geom_histogram(bins = 50) +
coord_cartesian(xlim = c(0, 100))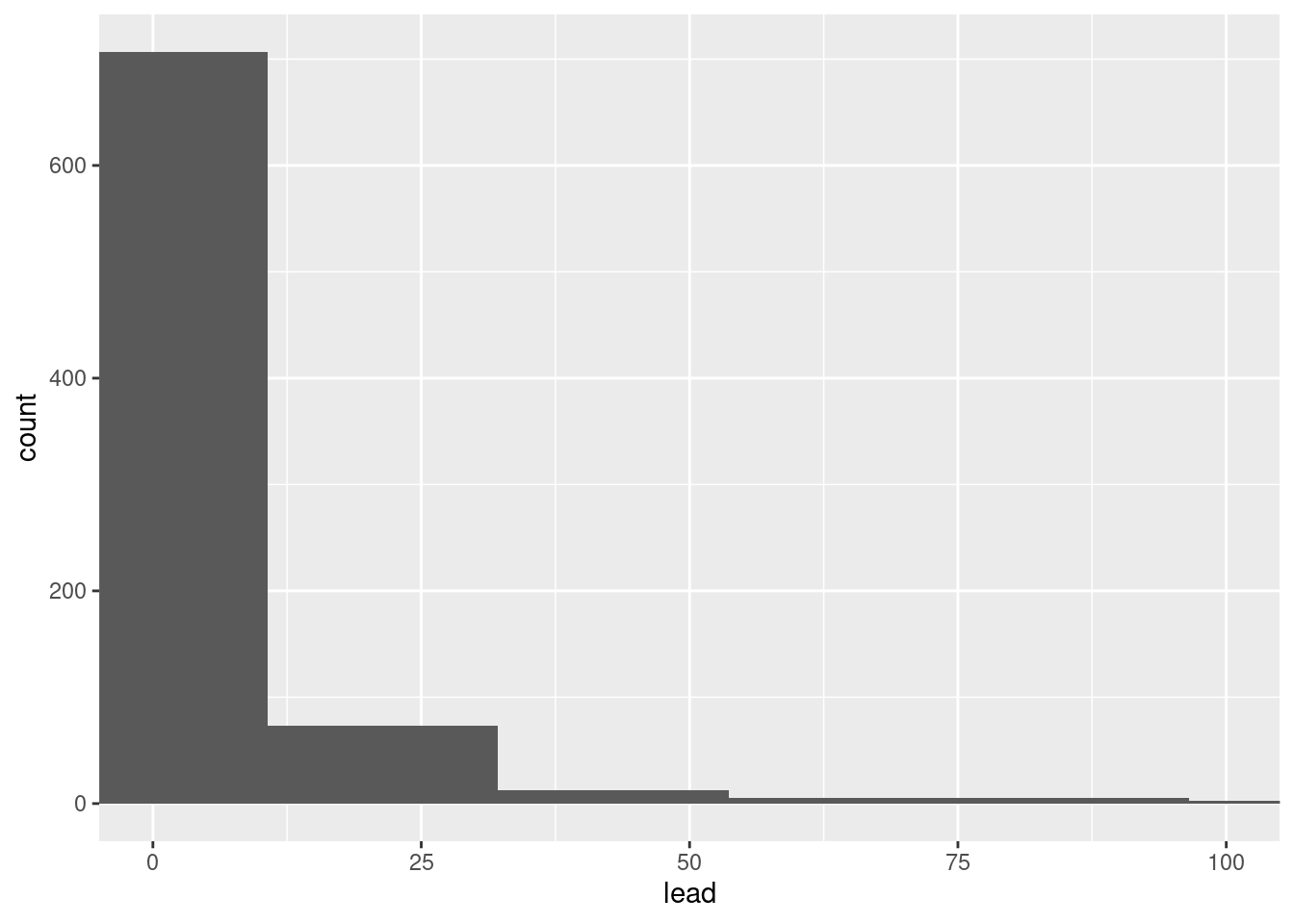
Let’s visualize some of our summary statistics on the plot.
Exercise 5: Add a new layer, geom_vline(xintercept = __, color = "red"), to the histogram below, filling in the blank with the mean.
mean_lead <- mean(flint$lead)
ggplot(flint, aes(x = lead)) +
geom_histogram(bins = 50) +
coord_cartesian(xlim = c(0, 100)) +
geom_vline(xintercept = summary_table$mean_lead, color = "red")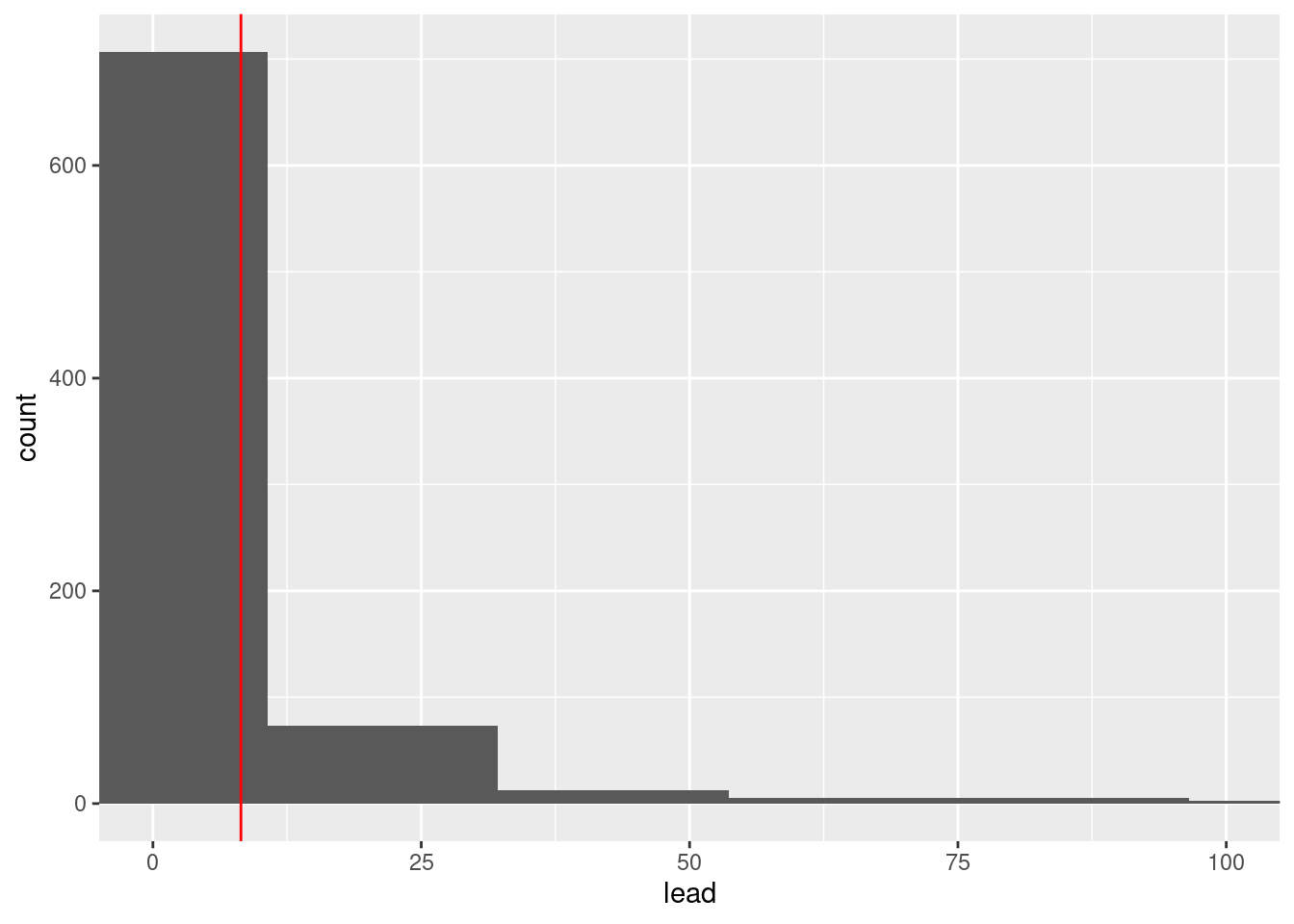
Exercise 6: Add one more layer which overlays the median, in a different color.
ggplot(flint, aes(x = lead)) +
geom_histogram(bins = 50) +
coord_cartesian(xlim = c(0, 100)) +
geom_vline(xintercept = summary_table$mean_lead, color = "red") +
geom_vline(xintercept = summary_table$median_lead, color = "blue")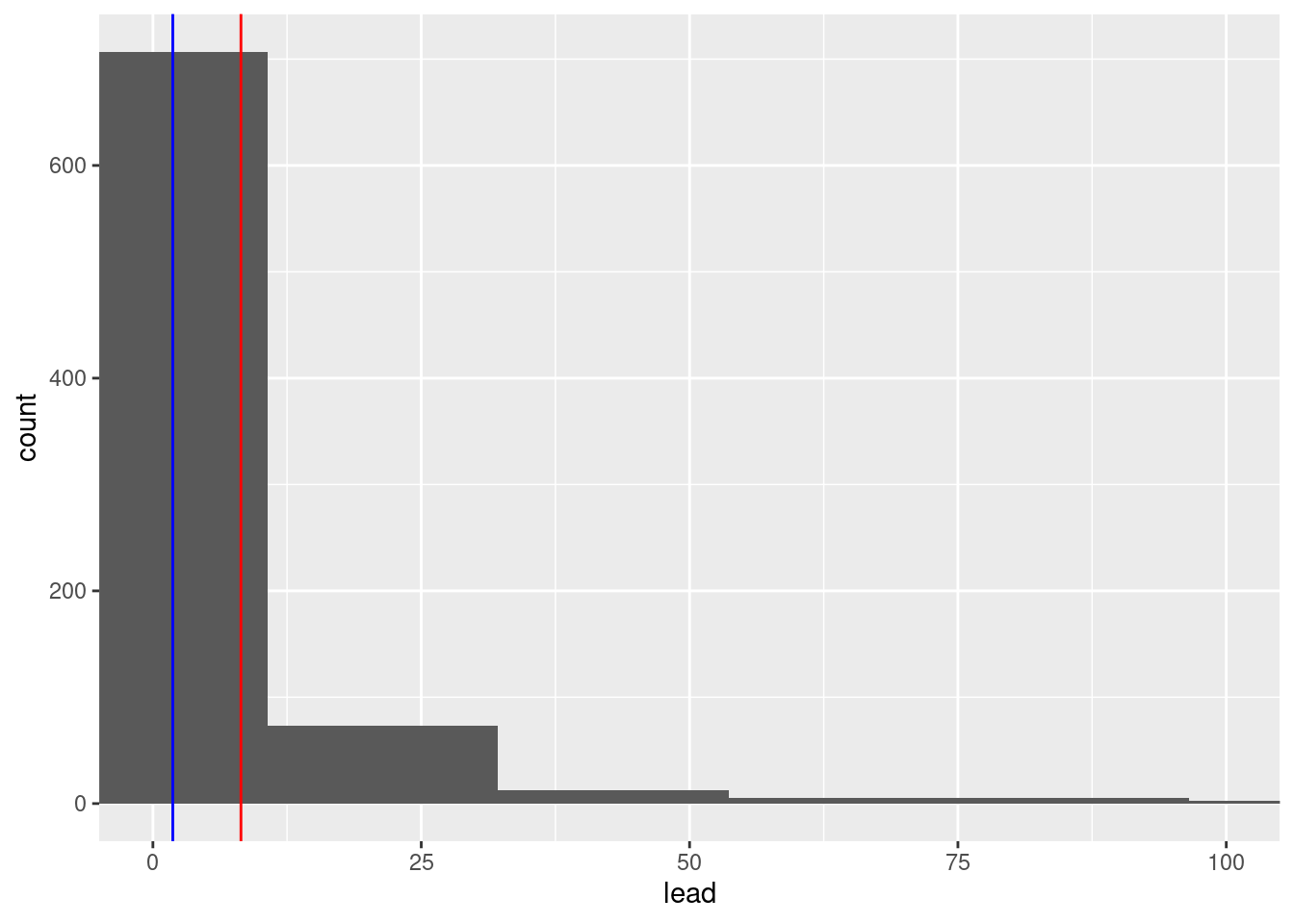
# add code hereNext, let’s narrow our focus to the zip codes 48503, 48504, 48505, 48506, and 48507 and observations with lead values less than 1,000 ppb.
flint_focus <- flint |>
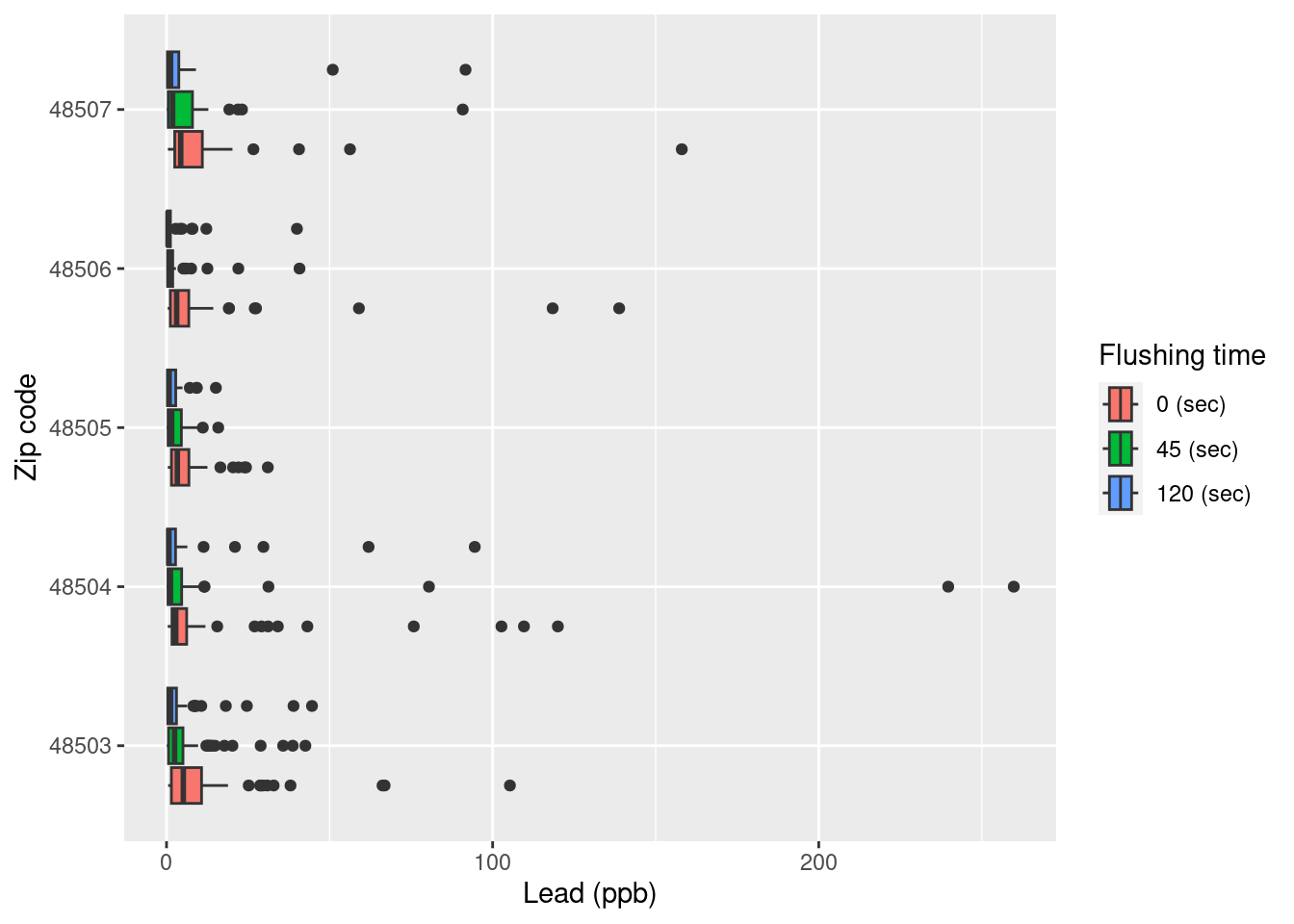
filter(zip %in% 48503:48507 & lead < 1000)Exercise 7: Below are side-by-side box plots for the three flushing times in each of the five zip codes we considered. Add x and y labels; add a title by inserting title = "title_name" inside the labs() function.
ggplot(data = flint_focus, aes(y = factor(zip), x = lead)) +
geom_boxplot(aes(fill = factor(draw))) +
labs(x = "Lead (ppb)", y = "Zip code", fill = "Flushing time") +
scale_fill_discrete(
breaks = c("first", "second", "third"),
labels = c("0 (sec)", "45 (sec)", "120 (sec)")
)
Exercise 8: Add labels for x, y, a title, and subtitle to the code below to update the corresponding plot.
ggplot(data = flint_focus, aes(y = factor(zip), x = lead)) +
geom_boxplot(aes(fill = factor(draw))) +
labs(
x = "Lead (ppb)", y = "Zip code", fill = "Flushing time",
title = "Lead amount by flushing time",
subtitle = "In five zip codes"
) +
scale_fill_discrete(
breaks = c("first", "second", "third"),
labels = c("0 (sec)", "45 (sec)", "120 (sec)")
) +
coord_cartesian(xlim = c(0, 50)) +
theme_bw()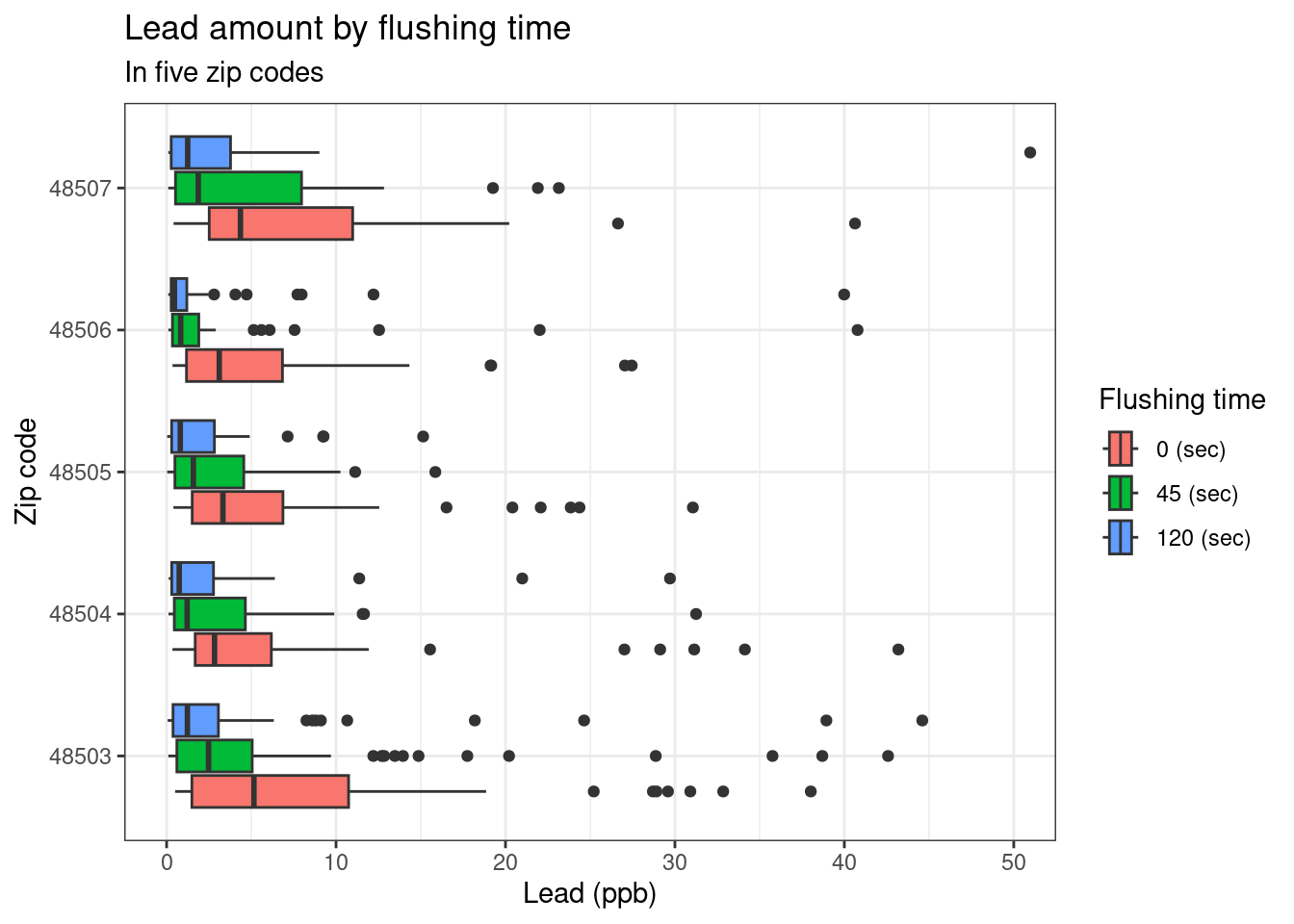
Exercise 9: What is the difference between the two plots? What are the advantages and disadvantages to each plot?
The first plot shows extreme outliers, while the second plot makes it easier to see the bulk of the distribution.
Langkjaer-Bain, R. (2017). The murky tale of Flint’s deceptive water data. Significance, 14: 16-21. doi: https://doi.org/10.1111/j.1740-9713.2017.01016.x.
Kelsey J. Pieper, et. al. Evaluating Water Lead Levels During the Flint Water Crisis. Edwards Environmental Science & Technology 2018 52 (15), 8124-8132 doi: https://doi.org/10.1021/acs.est.8b00791.